In-depth virological and immunological characterization of HIV-1 cure after CCR5Δ32/Δ32 allogeneic hematopoietic stem cell transplantation
Abstract
Despite scientific evidence originating from two patients published to date that CCR5Δ32/Δ32 hematopoietic stem cell transplantation (HSCT) can cure human immunodeficiency virus type 1 (HIV-1), the knowledge of immunological and virological correlates of cure is limited. Here we characterize a case of long-term HIV-1 remission of a 53-year-old male who was carefully monitored for more than 9 years after allogeneic CCR5Δ32/Δ32 HSCT performed for acute myeloid leukemia. Despite sporadic traces of HIV-1 DNA detected by droplet digital PCR and in situ hybridization assays in peripheral T cell subsets and tissue-derived samples, repeated ex vivo quantitative and in vivo outgrowth assays in humanized mice did not reveal replication-competent virus. Low levels of immune activation and waning HIV-1-specific humoral and cellular immune responses indicated a lack of ongoing antigen production. Four years after analytical treatment interruption, the absence of a viral rebound and the lack of immunological correlates of HIV-1 antigen persistence are strong evidence for HIV-1 cure after CCR5Δ32/Δ32 HSCT.
Main
Human immunodeficiency virus type 1 (HIV-1) persists in the body during antiretroviral therapy (ART) in latently infected CD4+ T cells, but allogeneic hematopoietic stem cell transplantation (HSCT) has been shown to substantially reduce the viral reservoir1,2. However, some of the reservoir-harboring immune cells are extremely long-lived3, partially resistant to chemotherapy regimens used during HSCT procedures and can cause viral rebound on analytical treatment interruption (ATI)4,5. Notably, both cases of successful HIV-1 cure published so far—the ‘London patient’ (IciStem no. 36) and the ‘Berlin patient’—received a CCR5Δ32/Δ32 allograft6,7 conferring extended resistance to HIV-1 due to the absence of surface expression of the CCR5 coreceptor.
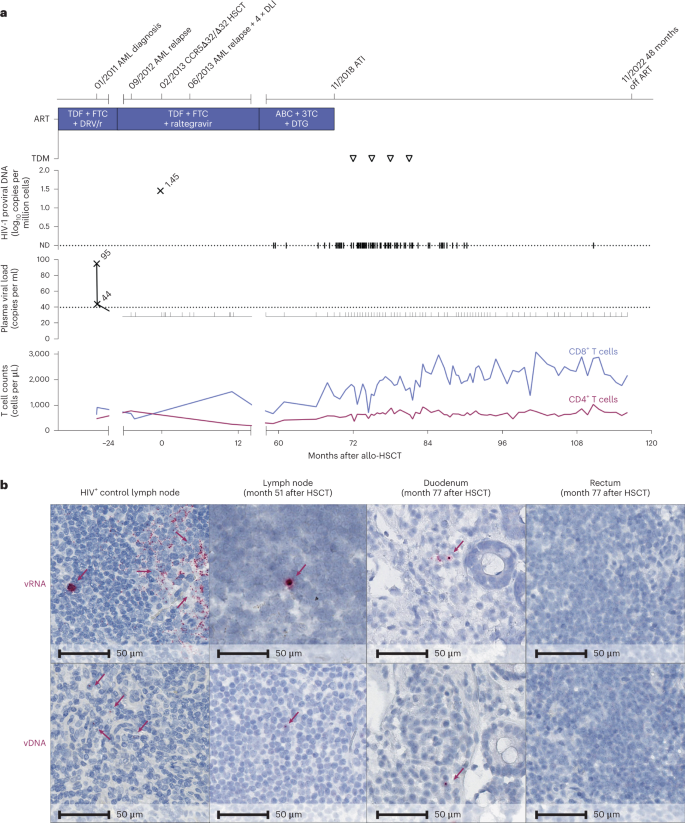
In this study, we provide a detailed longitudinal virological and in-depth immunological analysis of the peripheral blood and tissue compartments of a 53-year-old male patient (IciStem no. 19)8, alive and in good health 117 months after CCR5Δ32/Δ32 allogeneic HSCT and 48 months after ATI. The patient was diagnosed to be HIV-1 clade B positive in January 2008 and presented with a CD4+ T cell count of 964 cells per μl and an HIV-1 plasma viral load of 12,850 copies per ml (Centers for Disease Control and Prevention A1, which was no indication for initiation of ART according to the national guidelines at that time). In October 2010, an ART regimen with tenofovir disoproxil fumarate (TDF), emtricitabine (FTC) and darunavir and ritonavir (DRV/r) was initiated (503 CD4+ T cells per μl and 35,303 HIV-1 copies per ml), resulting in a continuously suppressed plasma viral load (Fig. 1a). In January 2011, the patient was diagnosed with acute myeloid leukemia (AML) M2 according to the French–American–British classification, which carried an inversion of chromosome 16 at p13q22, resulting in the CBFB–MYH11 fusion protein. The patient achieved hematological complete remission after chemotherapy, which included idarubicin, cytarabine, etoposide induction therapy and three high-dose cytarabine (HiDAC) consolidation cycles. To avoid drug–drug interactions, DRV/r was switched to raltegravir in March 2011.
a, Clinical events, ART regimen, HIV-1 plasma viral load, proviral load from PBMC and CD4+ and CD8+ counts. The triangles indicate negative results in the therapeutic drug monitoring (TDM) 3, 6, 9 and 12 months after ATI. ABC, abacavir; ND, not detectable. b, In situ hybridization assays for viral DNA (vDNA) and viral RNA (vRNA) revealed HIV-1 DNA and RNA traces in lymph node tissue (month 51) and duodenum but not rectum biopsies (month 77). Left panels: untreated HIV+ control patient.
In September 2012, the patient experienced an AML relapse but achieved a second complete remission after treatment with the A-HAM chemotherapy regimen (retinoic acid, HiDAC, mitoxantrone) and a second cycle of HiDAC. A systematic search identified a 10/10 HLA-matched (no mismatches in HLA-A, HLA-B, HLA-C, HLA-DR and HLA-DQ loci) unrelated female stem cell donor with a homozygous CCR5Δ32 mutation (Extended Data Table 1). After reduced-intensity conditioning with fludarabine, treosulfan and anti-thymocyte globulin, 8.74 × 106 unmodified CD34+ peripheral blood stem cells per kg of body weight were transplanted in February 2013. Immunosuppressive therapy consisted of cyclosporine and mycophenolate mofetil and was later changed to tacrolimus monotherapy. In June 2013, the patient experienced a second AML relapse. He achieved molecular oncological remission a third time after eight cycles with 5-azacytidine and four donor lymphocyte infusions (DLIs; 1 × 106, 10 × 106 and twice 50 × 106 donor T cells per kg of body weight). Thirty-four days after HSCT, full donor chimerism was established and retained except for a short period during the second relapse at months 3 and 4 after HSCT (Extended Data Fig. 1a). In 2014, elevated liver enzymes caused discussion about possible hepatic graft versus host disease (GvHD), but a liver biopsy in May 2015 was interpreted as drug-induced liver injury. In July 2014, the patient also experienced reactivation of cytomegalovirus (CMV) (duodenal ulcer) and herpes simplex virus 2 (genital ulcers and cerebral vasculitis) and human herpesvirus 8 and Epstein–Barr virus (viremia) but recovered after specific antiviral treatment of the CMV and herpes simplex virus 2 infections. After DLI, mild chronic GvHD of the eyes with bilateral keratoconjunctivitis sicca developed that persists until today. ART was continued throughout and proviral HIV-1 DNA and HIV-1 RNA remained undetectable despite intensified testing in clinical routine assays (Fig. 1a). However, multiple assessments of the HIV-1 viral reservoir in the peripheral blood and lymphoid and gut tissue before and after ATI revealed sporadic HIV-1 DNA traces at several time points, with a higher frequency compared to HIV-1-negative donors and no-template controls (Extended Data Table 2). Although rare, residual HIV-1 DNA and HIV-1 RNA were also detected by in situ hybridization (DNAscope and RNAscope assays) from histological sections of inguinal lymph node tissue from month 51 and some gut biopsies from month 77 (Fig. 1b); the number of HIV-1 RNA+ cells (2.61 ± 0.13 HIV-1 RNA+ cells per 105 cells) and HIV-1 DNA+ cells (5.08 ± 1.74 HIV-1 DNA+ cells per 105 cells) were only modestly above the limit of detection established for the assay. Importantly, neither HIV-1 p24, HIV-1 RNA or HIV-1 DNA was detectable in peripheral blood mononuclear cells (PBMCs) by repeated cell culture-based quantitative viral outgrowth assay nor by intact proviral DNA assay (Extended Data Table 2). Negative in vivo outgrowth assays using two different humanized mouse models confirmed the absence of replication-competent virus in the tested samples (Extended Data Fig. 2). Despite repeated in-depth reservoir assessments to determine whether or not the virus persisted and thus ensure the safest possible approach for ATI, the presence of residual replication-competent virus could not be completely ruled out7,9,10 and ATI ultimately remained the only way to prove HIV-1 cure8,9.
ART was discontinued 69 months after HSCT in November 2018, after careful consideration, and no antiretroviral agents were detected in four plasma samples collected after ATI (Fig. 1a). On cessation of ART, the patient remained without any clinical or laboratory signs of an acute retroviral syndrome. No rebound of plasma HIV-1 RNA occurred during ATI after 48 months in the absence of ART (Fig. 1a).
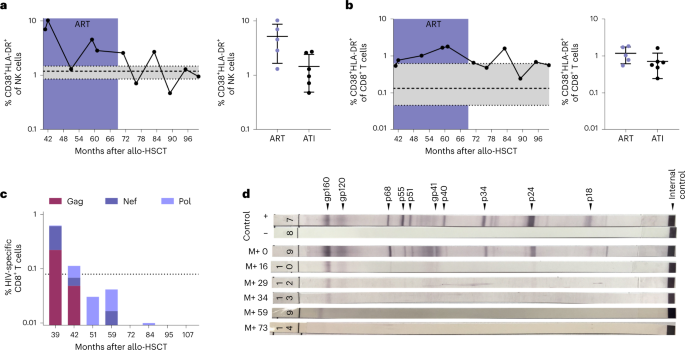
Extended immunological profiling before and after ATI demonstrated stable CD4+ T cell counts, absence of CCR5 expression (Extended Data Fig. 1c) and an immune status comparable to previous reports of people living with HIV (PLWH) after HSCT (with reduced naive T cell frequencies, elevated terminally differentiated effector memory T (TEMRA) cell frequencies and elevated frequencies of CD56− natural killer (NK) cells)8 (Extended Data Fig. 3). The activation levels of the patient’s peripheral blood NK cells and cytotoxic CD8+ T cells after ATI were within the range observed in HIV-1-negative controls (Fig. 2a,b; see also previously published data in ref. 8). Moreover, the immune cell composition and, in particular, the CD4+ T cell density was normal in the investigated lymphoid tissue at the time of sampling (month 51 after HSCT) and in gut tissue samples obtained after ATI (month 77 after HSCT; Extended Data Fig. 4a). Additionally, there was no evidence of elevated inflammation in the lymphoid and gut tissue (measured by MX1 expression) or gut barrier damage (measured by MPO expression) by immunohistochemical staining (Extended Data Fig. 4b,c).
a,b, NK cells (a) and CD8+ T cells (b) both showed decreasing and stable immune activation (coexpression of CD38 and HLA-DR). See also data from months 41–59 after HSCT for IciStem no. 19 as published in ref. 8. Data are shown as the mean with s.d. and a reference range from a healthy cohort (n = 8) in gray as the median with interquartile range (IQR). c, HIV-1-specific CD8+ T cell responses (production of IFNγ, TNFα, IL-2 and/or expression of CD107a) against HIV-1 Gag (burgundy), Nef (dark purple) and Pol (light purple) peptide pools waned after HSCT. See also the data from months 39 and 42 after HSCT for IciStem no. 19 as published in ref. 8. The dotted line represents the average background signal. d, Full-virus lysate immunoblot for antibodies against HIV-1 antigens revealed waning HIV-1-specific antibody responses after HSCT and prolonged weakening of gp160- and gp120-specific antibodies. M+, months after HSCT.
In month 39 after HSCT, HIV-1-specific CD8+ T cells were weakly detected on stimulation with overlapping peptide pools spanning HIV-1 Gag, Pol and Nef by intracellular cytokine staining (Fig. 2c; see also previously published data in ref. 8). However, the frequencies of HIV-1-specific T cells were substantially lower than previously observed for other PLWH8,10, further declined below threshold levels while still on ART and did not increase after ATI (Fig. 2c). Likewise, no significant T cell responses toward a set of 37 HIV-1 peptides known to be restricted by the patient’s major histocompatibility complex (MHC) class I molecules were detected before or after ATI as measured ex vivo by interferon-γ (IFNγ) enzyme-linked immunosorbent spot (ELISpot) (Supplementary Table 1) or MHC class I tetramer enrichment of an HLA-A*02-restricted reverse transcriptase epitope (RT-YV9) (Extended Data Fig. 5a). However, stimulation of PBMCs with the HLA-A*02-restricted RT-YV9 peptide and the gag-p6 peptide Gag07-121 elicited pronounced T cell responses in vitro after discontinuation of tacrolimus in month 56 that eventually decreased and were not detected in the most recent samples (Extended Data Fig. 5b). These short-term in vitro-expanded HIV-1-specific T cells were donor-derived because they carried the CCR5Δ32/Δ32 deletion (Extended Data Fig. 1b) suggesting antigen presentation to donor T cells in the peri-transplant period, despite continued ART8. In contrast, CMV-specific CD8+ T cells were strongly and consistently detected (Extended Data Fig. 5c), indicating that the waning of HIV-1-specific CD8+ T cells was probably due to the absence of stimulation by HIV antigens.
Immunoblot analyses of the HIV-1-specific antibody response paralleled the progressive loss of cellular HIV-1 reactivity: anti-gp120 and anti-gp160 antibodies showed the longest persistence (Fig. 2d), as reported previously2,7. At month 39 after HSCT, the peripheral blood HIV-1-specific antibody levels were below the cutoff for PLWH and comparable to HIV-1-negative individuals (Extended Data Fig. 5d). Because the maintenance of virus-specific responses is dependent on antigen exposure in chronic infection11, the extremely weak and waning HIV-1-specific T cell response and the decreasing levels of specific antibodies suggest that the HIV-1 reservoir capable of antigen production has been extremely depleted by the HSCT procedure and/or the graft versus HIV effect1,2.
Despite traces of HIV-1 DNA, there was no evidence of viral rebound or immunological correlates of antigen exposure. It is unclear whether the residual HIV-1 DNA signals stem exclusively from defective viral fragments or from an infinitesimally small pool of intact proviruses because proof for the absence of residual replication-competent virus was limited by the number of cells obtained and restricted accessibility of some anatomical compartments known to harbor the HIV-1 reservoir9,10. Therefore, it might be decisive that the patient was transplanted with a CCR5Δ32/Δ32 graft and that only an extremely low proportion (0.14%) of the proviral sequences retrieved before HSCT was characterized as possibly CXCR4-tropic, while potentially intact proviruses of the predominant R5-tropic population would not be able to propagate in this protective setting because of the absence of CCR5+ target cells4 (Extended Data Table 3).
The detailed observational characterization of single cases of HIV-1 cure after HSCT gives important insights but is anecdotal by nature and lacks the power of controlled prospective studies. Therefore, certain aspects of the cases published to date are of uncertain relevance for HIV-1 cure6,7, for example, underlying hematological malignancy, conditioning regimens or degree of GvHD as well as donor–recipient sex mismatch or the timing of ATI. However, two similarities are crucial for the outcome: all three patients achieved long-term remission after CCR5Δ32/Δ32 HSCT and harbored predominant R5-tropic virus strains. While CCR5Δ32/Δ32 HSCT cannot prevent the rebound of X4-tropic viruses5, this modification of the host’s immune system is a key component to prevent reservoir reseeding in PLWH with R5-tropic viruses requiring HSCT. Reservoir reduction during conditioning chemotherapy and immune-mediated HIV-1 clearance by donor cells (the ‘graft versus HIV effect’), and unspecific decay of latently infected cells driven by alloreactivity (GvHD; potentially further enhanced by DLI), might be additional, potential components that together led to the cure of HIV-1. However, it remains elusive to what extent reservoir depletion is necessary and contributes to HIV-1 cure in this and other cases2,6,7,8. One limitation of our study was the scarcity of samples available for detailed immunological and virological analysis directly before and after the HSCT.
Although HSCT using donors with a CCR5Δ32/Δ32 mutation is neither a low-risk nor an easily scalable procedure, its relevance to cure strategies is highlighted by recent reports of successful long-term HIV-1 remission after CCR5Δ32/Δ32-HSCT7,12,13. Expansion of this approach to introduce the CCR5Δ32 mutation into wild-type stem cell grafts using gene therapy in combination with new reservoir reduction strategies may hold the promise of an HIV-1 cure outside of life-threatening hematological malignancies. This third case of HIV-1 cure after allogeneic CCR5Δ32/Δ32 HSCT provides detailed information on the virological and immunological signature before and after ATI and generates valuable insights that will hopefully guide future cure strategies.
Methods
Ethics
The described individual (male, 53 years old as of 2022) was enrolled as patient no. 19 in the IciStem program at University Hospital Düsseldorf. ATI and examinations of the viral reservoir were performed after consultation of the ethics board of the Medical Faculty of the Heinrich Heine University Düsseldorf (official statement from 29 July 2016). Written informed consent was obtained from the patient for ATI and additionally performed immunological and virological studies (ethics board of the Medical Faculty of the Heinrich Heine University Düsseldorf no. 4261) in accordance with CAse REport (CARE) guidelines and the 2013 Declaration of Helsinki principles.
As controls for in situ hybridization assays and immunohistochemistry, a single lymph node and gut biopsies of one HIV-1-positive control (male, 56 years old) were collected in the ACS cohort, an ongoing, prospective multicenter acute HIV-1 infection cohort in Belgium, coordinated at the HIV Cure Research Center of Ghent University Hospital (ClinicalTrials.gov ID: NCT03449706) and approved by the ethics committee of Ghent University Hospital (no. BC-00812). The collection of rectum biopsies of one HIV-1-negative volunteer (male, 76 years old) undergoing screening colonoscopy at the HIV Cure Research Center at Ghent University Hospital was approved by the ethics committee of Ghent University Hospital (nos. BC-00812 and BC-11798). The lymph node of one HIV-1-negative individual (female, 41 years old) was provided by Knight BioLibrary (institutional review board-approved, no. IRB00004918) under full ethical approval by the Oregon Health & Science University institutional review board. Rectal tissue of one HIV-1-positive individual (male, 30 years old) for MPO staining was obtained from a study reported previously14.
For flow cytometry and MHC class I tetramer staining, eight HIV-1-negative controls (37.5% female, median age 25.5 years, range 21–28 years) and one HIV-1-positive control (male, 69 years old) were enrolled at the University Medical Center Hamburg-Eppendorf and were approved by the ethics board of the Hamburg Medical Association (nos. PV4780 and MC316/14).
All controls were enrolled on a voluntary basis, were not compensated and provided written informed consent, complying with the 2013 Declaration of Helsinki principles.
HIV-1 RNA quantification and ultrasensitive viral load
Quantification of HIV-1 RNA in plasma and cerebrospinal fluid samples was carried out with clinical routine assays and three different automated platforms during the observation period between January 2011 and October 2022. Until December 2014, the Abbott RealTime HIV-1 assay was used on the Abbott m2000 system. From January 2015 to July 2021, HIV-1 RNA quantification was performed on the Roche CAP/CTM using the COBAS AmpliPrep/COBAS TaqMan HIV-1 Test v.2.0. Since August 2021, quantification has been performed on the Roche c6800 system using the COBAS HIV-1 Test Kit. All analyses were carried out according to the manufacturer’s instructions with a detection limit of 40 copies per ml (Abbott) and 20 copies per ml (Roche).
Ultrasensitive residual viremia was measured in 9 ml of plasma after ultracentrifugation at 170,000g at 4 °C for 30 min, followed by viral RNA extraction using the m2000sp Abbott RealTime HIV-1 Assay and laboratory-defined application software from the instrument15. HIV-1 RNA was quantified with a validated in-house calibration curve set with a limit of detection of 0.56 copies per ml.
HIV-1 DNA quantification and droplet digital PCR
Clinical routine quantification of cellular HIV-1 DNA (proviral load) in PBMCs and buffy coat samples was performed with the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 Test v.2.0 on the Roche CAP/CTM platform or the Abbott RealTime HIV-1 Amplification Reagent Kit on the Abbott m2000rt Analyzer according to the manufacturer’s instructions. To exclude amplification of viral RNA during quantification of proviral DNA, cellular DNA was previously extracted automatically using the EZ1 DNA Blood 200 Kit (QIAGEN) via the EZ1 Advanced XL Platform or with the Abbott mSample-Preparation System Kit on the Abbott Tecan m2000sp platform and used in equal amounts for quantification. The normalization of the detected HIV-1 DNA copies to the number of PBMCs was carried out by quantifying the housekeeping genes β-actin or β-globin.
Droplet digital PCR (ddPCR) was performed from different samples: PBMCs were isolated from blood samples. From there, CD4+ T bulk cells and four different subsets (naive T, central memory T, transitional memory T and effector memory T cells) were isolated by cell sorting. Gut biopsies (ileum, rectum and duodenum) were disaggregated by dithiothreitol/EDTA-based treatment for epithelial layer removal followed by overnight nonenzymatic disruption of the tissue to obtain lamina propria cell suspension16. CD45+ cells were subsequently purified by flow sorting using the BD FACS Aria cytometer (BD Biosciences). Follicular helper CD4+ memory T cells (CD3+CD4+CD45RA+PD1+CXCR5+) were sorted by flow cytometry from lymph node biopsy specimens using the BD FACS Aria cytometer.
In all cases, the isolated cells were lysed and viral DNA was quantified with ddPCR with HIV-1 LTR and Gag primers, as indicated in Supplementary Table 2 and as described previously16,17,18. Undetectable values are represented as a specific limit of detection adjusted to the number of cells used in each sample.
Quantification of intact provirus was assessed using lysed extracts of CD4+ T cells. Duplex ddPCR was performed using the packaging signal (Ψ) and nonhypermutated envelope gene (Env) primer and probe sets, according to the original proviral DNA assay protocol19.
Viral coreceptor tropism analysis
Using PBMCs from day −7 before HSCT, the proviral load was determined and the Env region containing the complete V3 loop was amplified using an in-house protocol20 with primers as indicated in Supplementary Table 2. Genotypic coreceptor usage was predicted using the Geno2pheno [coreceptor] tool21 after Sanger sequencing.
Additionally, the amplified product was deep-sequenced using the Illumina MiSeq platform22. The sequence data are available via the NCBI GenBank database (accession nos. OP712709–OP713600). Genotypic coreceptor usage for all 17,701 reads (535 different V3 amino acid sequence variants) covering the whole V3 region was predicted using the Geno2pheno [454] tool23. In detail, for each variant, the false positive rate (FPR) of falsely classifying an R5-tropic virus as an X4-tropic virus was predicted; according to the determined cutoff of ≤3.5% FPR the single sequences were defined as R5- or X4-tropic. The total sample was considered to be X4-tropic if ≥2% of the viral population was scored as X4-tropic24. Of all 535 detected V3 variants, nine were selected for the phenotypic assay considering the following conditions to cover all variations: predicted R5, high FPR (nos.1–3); predicted R5, high FPR, high percentage (no. 4); predicted R5, low FPR, highest percentage (no. 5); predicted R5, low FPR (no. 6); predicted X4, low FPR, highest percentage (no. 7); predicted X4, low FPR (no. 8); predicted X4, lowest FPR (no. 9).
These nine sequences, which encode the full V3 region from nucleotide 7,110 to 7,217 (based on the HxB2 reference sequence), were cloned into the pHXB2-Δgp120-V3 vector25. The same vectors carrying either the gp120-V3 sequence of HxB2 (X4-tropic) or of BaL (R5-tropic) served as positive controls. The chimeric viruses were tested in cell culture with MT2 cells and the U373-MAGI-CCR5E (CD4+CCR5+CXCR4−) and U373-MAGI-CXCR4CEM (CD4+CCR5−CXCR4+) cell lines, all obtained from the HIV reagents program.
Antiretroviral drug screening in plasma
Quantification of the following antiretroviral drug concentrations was performed using a liquid chromatography–tandem mass spectrometry method (4–5) validated according to Food and Drug Administration and European Medicines Agency guidelines on the TSQ Quantiva UltiMate 3000 RSLC: FTC, lamivudine (3TC), tenofovir, efavirenz, etravirine, nevirapine, rilpivirine, amprenavir, atazanavir, darunavir, indinavir, lopinavir, nelfinavir, ritonavir, saquinavir, tipranavir, dolutegravir (DTG), elvitegravir, raltegravir and maraviroc. Testing was performed by the Clinical Pharmacology Department of the University Medical Center in Utrecht at months 72, 75, 78 and 81 after HSCT (months 3, 6, 9 and 12 after ATI, respectively).
HIV-1 clade B in situ hybridization (RNAscope/DNAscope)
HIV-1 in situ hybridization was performed as described previously with a set of HIV-1 clade B lineage-specific in situ hybridization riboprobes (cat. nos. 416111 and 425531, ACDBio) as described previously26. The target probes used bind to all HIV-1 genes and cover just over 4.2 kb of the genome. However, the assay cannot distinguish infected cells with intact HIV-1 genomes from cells with defective HIV-1 genomes. The FPR for this study was determined in two ways: First, using clinical specimens from IciStem no. 19, the HIV-1-specific probe was replaced with assay buffer while all other assay conditions remained constant. Second, the full assay, including the HIV-1-specific probe, was performed on an HIV-1-uninfected cell line control. The highest FPR across all controls determined the specific threshold (logarithm of the odds = 1.83 HIV-1 RNA+ cells per 105 cells and 4.18 HIV-1 DNA+ cells per 105 cells) for the assay.
Immunohistochemistry
Immunohistochemistry was performed as described previously26. A multi-staining approach of CD4/CD68/CD163 to quantify CD4+ T cells was performed and allowed the intense staining of the macrophage/myeloid cell markers to mask the faint CD4 expressed on these cells and to distinctly identify CD4+ T cells from myeloid lineage cells as described previously. Heat-induced epitope retrieval was performed by heating sections in 0.01% citraconic anhydride containing 0.05% Tween-20 for Mx1 or citrate pH 6.0 for MPO in a pressure cooker (Biocare Medical) set at 122 °C or 125 °C for 30 s. Slides were incubated with blocking buffer and then with mouse anti-Mx1 monoclonal antibody (clone M143, 1:2,000; a gift from G. Kochs and the Department of Virology of the University of Freiburg) diluted in blocking buffer for 1 h at room temperature with or with rabbit anti-MPO (1:5,000, cat. no. A0398, Dako) diluted in blocking buffer overnight at 4 °C. After washing, endogenous peroxidases were blocked using 1.5% (v/v) H2O2 in Tri-buffered saline, pH 7.4, and the slides were incubated with rabbit or mouse Polink (−1 or −2) horseradish peroxidase and developed with ImmPACT 3,3′-diaminobenzidine (Vector Laboratories), according to the manufacturer’s recommendations. All slides were washed in tap water, counterstained with hematoxylin, mounted in Permount (Thermo Fisher Scientific) and scanned at high magnification (200×) using the AT2 Aperio System (Leica); all high-resolution images were stored in HALO (Indica Labs). Representative regions of interest were identified and high-resolution images were extracted from these whole-tissue scans using the HALO figure generation tool.
Quantitative viral outgrowth assays
In months 36 and 37, 2 × 107 PBMCs were isolated from whole-blood samples each and CD8+ T cells were removed (CD8-pluriBead). Activated PBMCs (phytohemagglutinin (PHA) and IL-2) were cultivated in bulk cultures directly or in co-culture with PBMCs (CD8+ T cell-depleted) of HIV-1-negative donors or in co-culture with TZM-bl HIV reporter cells (National Institutes of Health (NIH) HIV reagents program) for 4 weeks. Weekly, cell culture supernatants were tested using a sensitive reverse transcriptase assay (CAVIDI) and cellular DNA was analyzed by viral DNA PCR and lysates of TZM-bl cells for luciferase activity.
Leukapheresis was obtained 39 and 80 months after stem cell transplantation to measure infectious units per million CD4+ T cells. We set up a limiting dilution virus culture assay using the maximum cell number available (23 and 203 × 106 CD4+ T cells at months 39 and 80, respectively). A limiting dilution virus culture assay was used to measure the replication-competent reservoir in purified CD4+ T cells as described previously27 with the minor modification of using three donors with three different activators (PHA 5 μg ml−1, PHA 0.5 μg ml−1 and OKT3) as blast cells to boost the virus. The frequency of infectious HIV-1 units per million CD4+ T cells was calculated based on the 14-d p24Gag readout and using IUPMStats v.1.0, based on the maximum likelihood method (http://silicianolab.johnshopkins.edu/).
In vivo outgrowth assays in humanized mice
As an in vivo measure of residual replication-competent reservoir cells in the blood, we used two different humanized mouse models to transfer patient-derived CD4+ T cells, both at month 42 (refs. 28,29).
For the BALB/c Rag2−/−γc−/− mice, the experimental protocols were performed according to the guidelines of the German Animal Protection Law and reviewed and approved by the Hamburg Medical Association (nos. OB-050/07 and WF-010/2011) and the Hamburg Free and Hanseatic City, Authority for Health and Consumer Protection (nos. 63/09 and 23/11). Mice were kept on a 12/12-h dark–light cycle (lights on at 6:00) in a temperature-controlled (21–23 °C) and humidity-controlled (45–65%) environment. Mice were transplanted with CD8+-depleted PBMCs according to ref. 29. For this, 6-week-old male or female mice were preconditioned with an intraperitoneal injection of 60–80 µl of Clophosome-A Clodronate Liposomes (FormuMax Scientific). Forty-eight hours later, mice were irradiated with a dose of 4 Gy (at 4 h before transplantation) from a cesium 137 source at 3.75 Gy min−1 (CSL-12; Conservatome). Subsequently, mice were transplanted with 1 × 106 cells in 150 μl PBS containing 0.1% human AB serum (PAN Biotech) by intraperitoneal injection. At 7 weeks after transplantation, mice were subjected to end analyses. Human cell engraftment was verified by fluorescence-activated cell sorting (FACS) analysis of peripheral blood samples using retro-orbital sampling. Likewise, viremia was assayed by diluting cell-free mouse plasma with human serum (PAN Biotech) using the ultrasensitive (<20 HIV-1 RNA copies per ml) COBAS AmpliPrep/Cobas TaqMan HIV-1 Test v.2.0 (Hoffmann-La Roche). For the analysis of peripheral cells, 50–100 μl of blood was collected from the retro-orbital venous sinus into 100 μl of bleeding buffer (PBS plus 10 mM EDTA) and red blood cells were lysed by treatment with Red Blood Cell Lysing Buffer (Sigma-Aldrich). The white blood cell pellet was resuspended in FACS buffer (PBS containing 2% FCS and 2 mM EDTA) and stained with monoclonal antibodies. Single-cell suspensions of bone marrow, spleen and liver were prepared at necropsy by manual tissue dissection and filtering through a sterile 70-μm nylon mesh (BD Biosciences) for antibody staining and FACS analysis. Stained cells were analyzed in FACS buffer plus 1% paraformaldehyde using a FACSCanto (BD Biosciences) system with the BD FACSDiva software v.5.0.3 and FlowJo software v.7/9 (FlowJo LLC). To monitor human cell engraftment, retro-orbitally collected cells were stained with monoclonal antibodies raised against mouse CD45.2 (104), human CD45 (H130), CD4 (RPA-T4) and human CD3 (UCHT1) (eBioscience). Isotype antibodies and cells obtained from non-transplanted mice served as negative staining controls.
For NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice, procedures were performed according to protocol 8927, which was reviewed by the Animal Experimentation Ethics Committee of the University Hospital Germans Trias i Pujol (registered as B9900005) and approved by the Catalan government according to current national and European Union legislation on the protection of experimental animals. Mice were kept on a 12/12-h dark–light cycle (lights on at 8:00) in a temperature-controlled (20–24 °C) and humidity-controlled (40–70%) environment. Mice balanced in sex were supervised daily according to a strict protocol to ensure their welfare and were euthanized, if required, with isoflurane (inhalation excess). Experiments were followed as published previously2,28. Briefly, 80 million purified CD4+ T cells were infused in five 7-week-old mice (16 million per mouse). Whole-blood samples were collected at weeks 2, 4 and 6 when mice were euthanized. At every time point, plasma was used for the quantification of HIV-1 RNA. Whole blood was stained to calculate the human cell engraftment as the percentage of human CD45+ cells in the blood. Whole ACK-lysed blood cells and mechanically disaggregated murine spleen biopsies were used for HIV-1 DNA quantification by ddPCR with HIV-1 LTR and gag primers as described above.
Immunophenotyping
All analyses were performed with thawed PBMCs as described previously8. A total of 1–2 × 106 cells were used for the flow cytometry analyses and stained with Zombie NIR fixable viability dye (BioLegend) according to the manufacturer’s instructions and one of the following antibody panels. For the analysis of conventional T cells, fluorochrome-conjugated monoclonal antibodies targeting CCR5 (clone 2D7, BUV737, BD Biosciences), CD8 (clone RPA-T8, BV785), HLA-DR (clone L243, BV711), CD45RA (clone HI100, BV650), CXCR4 (clone 12G5, BV605), CCR7 (clone G043H7, BV421), CD4 (clone SK3, PerCP-Cy5.5), CD27 (clone M-T271, FITC, BD Biosciences), CD38 (clone HB-7, PE-Cy7), PD-1 (clone EH12.2H7, PE-Dazzle), CD25 (clone M-A251, PE), CD3 (clone UCHT1, Alexa Fluor 700), CD127 (clone A019D5, Alexa Fluor 647), CD14 (clone 63D3, APC-Cy7) and CD19 (clone HIB19, APC-Cy7). For the analysis of unconventional T cells and NK cells, fluorochrome-conjugated monoclonal antibodies targeting CD56 (clone NCAM16.2, BUV737, BD Biosciences), CD8 (clone RPA-T8, BV785), HLA-DR (clone L243, BV711), CD16 (clone 3G8, BV650), TCR Vα7.2 (clone 3C10, BV605), NKG2D (clone 1D11, BV421), CD4 (clone SK3, PerCP-Cy5.5), TCR Vδ2 (clone B6, FITC), CD38 (clone HB-7, PE-Cy7), CD161 (clone HP-3G10, PE-Dazzle), TCR γδ (clone 11F2, PE, BD Biosciences), CD3 (clone UCHT1, Alexa Fluor 700), CD127 (clone A019D5, Alexa Fluor 647), CD14 (clone 63D3, APC-Cy7) and CD19 (clone HIB19, APC-Cy7). Unless stated otherwise, antibodies were purchased from BioLegend. Before acquisition, cells were fixed with paraformaldehyde. All samples were acquired on an LSRFortessa analyzer (BD Biosciences) run by the FACSDiva software v.8 (BD Biosciences). Analysis of the flow cytometry data was performed in FlowJo v.10.7 for Windows. The gating strategy is depicted in Supplementary Fig. 2.
Intracellular cytokine staining
T cell stimulation and intracellular cytokine staining were performed as described previously8. Briefly, thawed PBMCs were rested overnight at 37 °C and 5% CO2 in RPMI medium (RPMI 1640 supplemented with L-glutamine and antibiotics) with 20% heat-inactivated FCS. Cells were then incubated with overlapping peptide pools encompassing HIV-1 consensus subtype B Gag, Pol and Nef or HCMV pp65 (all obtained through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH, cat. nos. 12425, 12438, 12545 and 11549) at 2 μg ml−1 and anti-CD28/anti-CD49d costimulation (BD Bioscience) at 1 μl ml−1. No peptides were added to the negative controls. Phorbol myristate acetate (80 ng ml−1) together with ionomycin (1 μg ml−1; Sigma-Aldrich) was used as a positive control. Anti-CD107a V450 (BD Bioscience) was added to all conditions. Golgi stop (1 μg ml−1; BD Biosciences) and brefeldin A (10 μg ml−1; Sigma-Aldrich) were added 30 min after the start of all incubations. Cells were then stained with the LIVE/DEAD Fixable Aqua Dead Cell Stain kit (Thermo Fisher Scientific) and with anti-CD3 Alexa Fluor 700, anti-CD4 APC and anti-CD8a APC-Cy7 antibodies (BD Biosciences). Cytofix and Cytoperm (BD Biosciences) were used for cell permeabilization before staining for intracellular markers. Intracellular staining used anti-IFNγ PE-Cy7, IL-2 FITC and anti-tumor necrosis factor-α (TNFα) PE-CF594 (BD Biosciences). Cell staining was then measured with an LSRII flow cytometer (BD Biosciences). Results were analyzed with FlowJo v.10.5. Because of the limited number of circulating T cells at some time points after allo-HSCT, results were only considered when at least 1,000 CD8+ T cells could be analyzed and the number of positive events was at least 50% higher than the negative control. The background signal of the negative control was subtracted from the signal obtained with the peptide pools. The gating strategy is depicted in Supplementary Fig. 3.
MHC class I tetramer staining
MHC class I tetramer staining was performed as described previously30 using PBMCs from months 51, 61, 75 and 87 after HSCT. Thawed PBMCs were stained with APC-A*02:01 HIV-1 RT-YV9 (YQYMDDLYV) and additionally BV421-A*02:01 CMV pp65 (NLVPMVATV) in the unenriched fractions, both obtained from the NIH Tetramer Core Facility. Tetramer enrichment was performed with anti-APC microbeads applying magnetic-activated cell sorting technology (Miltenyi Biotec) according to the manufacturer’s protocol. Cells were stained with Zombie NIR fixable viability dye (BioLegend) according to the manufacturer’s instructions and fluorochrome-conjugated monoclonal antibody targeting CD45RA (clone HI100, BUV737, BD Biosciences), CD38 (clone HB-7, BUV395, BD Biosciences), Tim-3 (clone F38-2E2, BV785), CD8 (clone RPA-T8, BV711), PD-1 (clone EH12.2H7, BV650), TIGIT (clone A15153G, BV605), CD3 (clone UCHT1, BV510), CD39 (clone A1, BV421; only enriched fractions), CD127 (clone A019D5, PerCP-Cy5.5), CD27 (clone M-T271, FITC, BD Biosciences), CCR7 (clone G043H7, PE-Cy7), HLA-DR (clone L243, Alexa Fluor 700), CD14 (clone 63D3, APC-Cy7) and CD19 (clone HIB19, APC-Cy7). Before acquisition, cells were fixed with paraformaldehyde. All samples were acquired on an LSRFortessa analyzer (BD Biosciences) run by the FACSDiva software v.8 (BD Biosciences). The analysis of the flow cytometry data was performed in FlowJo v.10.7 for Windows.
Ex vivo detection of HIV-1-specific T cells by IFNγ ELISpot
In the IFNγ ELISpot assay, 2 × 105 PBMCs per well were incubated for 40 h with HIV-1 peptides corresponding to known cytotoxic T lymphocyte (CTL) epitopes restricted by the patient’s HLA alleles (Supplementary Table 1) at a final concentration of 6 μg ml−1. Concanavalin A and PHA (Sigma-Aldrich) served as positive controls. Peptides were either purchased from EMC Microcollections or kindly provided by the NIH AIDS Research and Reference Reagent Program or by the AIDS Reagent Project of the Medical Research Council and the European Community EVA Programme.
In vitro expansion and analysis of HIV-1-specific T cells
Five million PBMCs each were stimulated with peptides corresponding to known CTL epitopes restricted by the patient’s HLA alleles at a final concentration of 6 μg ml−1 in 1 ml of RPMI 1640 medium (Gibco) containing 10% heat-inactivated FCS (Gibco), 1% L-glutamine (2 mmol l−1), penicillin (100 U ml−1), streptomycin (100 μg ml−1), HEPES (10 mmol l−1) (Merck) and 10 U ml−1 recombinant IL-2 (Proleukin). After 5–32 d, outgrowing cells were analyzed by IFNγ ELISpot assay for recognition of the peptides at a cell concentration of 1 × 105 cells per well. ELISpot assays were conducted using R5AB medium consisting of RPMI 1640 medium with supplements and 5% heat-inactivated human AB serum (Sigma-Aldrich).
Detection of CCR5-32-bp deletion
Cellular DNA was isolated from PBMC and T cell expansions using the NucleoSpin Blood QuickPure kit (Macherey-Nagel) according to the manufacturer’s instructions. Aliquots of DNA were amplified using Taq polymerase (Invitrogen) and 40 cycles (94 °C, 40 s; 56 °C, 50 s; 72 °C, 60 s) in a volume of 25 μl using the primers CCR5-5 760 (5′-CTGCAGCTCTCATTTTCC-3′) and CCR5-3 982 (5′-TCAGGAGAAGGACAATGTTG-3′). By separating the reaction products (8 μl) on 2% metaphore agarose (FMC) with MIDORI green, the presence of the 32-bp deletion could be visualized. As an additional control for the method, the visualized amplification product of the RT-YV9-specific CTL line was cut out from the agarose gel and analyzed with Dye Terminator Cycle Sequencing (PE, Applied Biosystems). The PCR products were sequenced on both strands using the same oligonucleotides as primers. PBMC and B cell lines from CCR5 wild-type, Δ32 heterozygous and Δ32 homozygous individuals served as controls.
Quantification of anti-HIV-1 antibodies
Specific anti-HIV-1 antibodies in plasma samples were measured using a qualitative full-virus lysate immunoblot assay (New LAV Blot, Bio-Rad Laboratories) based on an indirect enzyme-linked immunosorbent assay technique. Ready-to-use nitrocellulose strips contained all inactivated HIV-1 constituent proteins separated by their molecular weights and an internal anti-IgG control. In contrast to recombinant immunoblots, the position of the antigen bands can vary between lots. Additionally, the quantitative standard VITROS anti-HIV-1 immunoassay, a low-sensitivity version of the VITROS anti-HIV-1 immunoassay (Ortho Clinical Diagnostics), and the limiting antigen-avidity assay were performed as described previously31 to assess anti-HIV-1 antibody levels.
Statistics and reproducibility
Graphs were created and statistical analyses were performed using Prism 7 for Windows (GraphPad Software). For statistical testing, a nonparametric two-tailed Mann–Whitney U-test was performed. Reference values from healthy controls are displayed as the median with IQR. Sex bias could not be considered because this study focused on one specific individual.
Biological replicates (that is, samples at different time points) were measured in all experiments except in situ hybridization, immunohistochemistry and CCR5-PCR given the limited clinical material, tropism analysis given lack of viral sequences due to undetectable (pro)viral loads and mVOAs given ethical concerns and limited clinical material. Technical replicates (that is, repeated measuring of the same sample) were measured as follows: for clinical viral load testing at each time point, two separate plasma samples were collected and measured independently from one another with similar results; for ddPCR, technical replicates were measured as indicated in Extended Data Table 2 with similar results (any divergent results are reported due to extremely rare abundance of HIV-1 DNA traces); for mVOAs, n = 2 and n = 5 mice were transplanted with the tested cells and generated similar results; the in situ hybridization, immunohistochemistry and flow cytometry experiments were not technically replicated given the limited clinical material; CCR5-PCR was done from PBMCs ex vivo and two different T cell expansions with similar results; most HIV-1-specific antibody immunoblots were measured once due to limited clinical material. The conclusiveness comes from coherent results due to periodic measurements.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Figures 1 and 2 and Extended Data Figs. 1–5 contain raw data. The HIV-1 sequence data are available via the NCBI GenBank database (accession nos. OP712709–OP713600). The other data that support the findings of this study are available upon request from the corresponding authors B.-E.O.J. and J.S.z.W. within the data protection constraints of the written informed consent signed by the study participants (that is, pseudonymized data only); they will be made available within 6 weeks. The data are not publicly available because they contain information that could compromise the participants’ privacy. Source data are provided with this paper.